Lorentz-Equivariant GNN
Contents
Lorentz-Equivariant GNN#
Now we will look at graph neural networks using the PyTorch Geometric library: https://pytorch-geometric.readthedocs.io/. See [2] for more details.
# For Colab
!pip install torch_geometric
!pip install torch_sparse
!pip install torch_scatter
Requirement already satisfied: torch_geometric in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (2.1.0)
Requirement already satisfied: requests in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from torch_geometric) (2.28.1)
Requirement already satisfied: jinja2 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from torch_geometric) (3.1.2)
Requirement already satisfied: scipy in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from torch_geometric) (1.7.3)
Requirement already satisfied: tqdm in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from torch_geometric) (4.64.1)
Requirement already satisfied: pyparsing in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from torch_geometric) (3.0.9)
Requirement already satisfied: numpy in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from torch_geometric) (1.21.6)
Requirement already satisfied: scikit-learn in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from torch_geometric) (1.0.2)
Requirement already satisfied: MarkupSafe>=2.0 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from jinja2->torch_geometric) (2.1.1)
Requirement already satisfied: charset-normalizer<3,>=2 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from requests->torch_geometric) (2.1.1)
Requirement already satisfied: certifi>=2017.4.17 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from requests->torch_geometric) (2022.9.24)
Requirement already satisfied: idna<4,>=2.5 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from requests->torch_geometric) (3.4)
Requirement already satisfied: urllib3<1.27,>=1.21.1 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from requests->torch_geometric) (1.26.12)
Requirement already satisfied: threadpoolctl>=2.0.0 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from scikit-learn->torch_geometric) (3.1.0)
Requirement already satisfied: joblib>=0.11 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from scikit-learn->torch_geometric) (1.2.0)
Requirement already satisfied: torch_sparse in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (0.6.15)
Requirement already satisfied: scipy in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from torch_sparse) (1.7.3)
Requirement already satisfied: numpy<1.23.0,>=1.16.5 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from scipy->torch_sparse) (1.21.6)
Requirement already satisfied: torch_scatter in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (2.0.9)
import torch
import torch_geometric
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
from tqdm.notebook import tqdm
import numpy as np
local = False
# For Colab
!pip install wget
import wget
!pip install -U PyYAML
!pip install uproot
!pip install awkward
!pip install mplhep
Requirement already satisfied: wget in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (3.2)
Requirement already satisfied: PyYAML in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (6.0)
Requirement already satisfied: uproot in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (4.3.7)
Requirement already satisfied: numpy in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from uproot) (1.21.6)
Requirement already satisfied: setuptools in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from uproot) (65.5.1)
Requirement already satisfied: awkward in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (1.10.1)
Requirement already satisfied: numpy>=1.13.1 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from awkward) (1.21.6)
Requirement already satisfied: importlib-resources in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from awkward) (5.10.0)
Requirement already satisfied: packaging in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from awkward) (21.3)
Requirement already satisfied: zipp>=3.1.0 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from importlib-resources->awkward) (3.10.0)
Requirement already satisfied: pyparsing!=3.0.5,>=2.0.2 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from packaging->awkward) (3.0.9)
Requirement already satisfied: mplhep in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (0.3.26)
Requirement already satisfied: mplhep-data in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from mplhep) (0.0.3)
Requirement already satisfied: packaging in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from mplhep) (21.3)
Requirement already satisfied: matplotlib>=3.4 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from mplhep) (3.5.3)
Requirement already satisfied: uhi>=0.2.0 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from mplhep) (0.3.2)
Requirement already satisfied: numpy>=1.16.0 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from mplhep) (1.21.6)
Requirement already satisfied: pyparsing>=2.2.1 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from matplotlib>=3.4->mplhep) (3.0.9)
Requirement already satisfied: pillow>=6.2.0 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from matplotlib>=3.4->mplhep) (9.3.0)
Requirement already satisfied: cycler>=0.10 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from matplotlib>=3.4->mplhep) (0.11.0)
Requirement already satisfied: python-dateutil>=2.7 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from matplotlib>=3.4->mplhep) (2.8.2)
Requirement already satisfied: fonttools>=4.22.0 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from matplotlib>=3.4->mplhep) (4.38.0)
Requirement already satisfied: kiwisolver>=1.0.1 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from matplotlib>=3.4->mplhep) (1.4.4)
Requirement already satisfied: typing-extensions>=3.7 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from uhi>=0.2.0->mplhep) (4.4.0)
Requirement already satisfied: six>=1.5 in /opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages (from python-dateutil>=2.7->matplotlib>=3.4->mplhep) (1.16.0)
import yaml
import os.path
# WGET for colab
if not os.path.exists("definitions_lorentz.yml"):
url = "https://raw.githubusercontent.com/jmduarte/iaifi-summer-school/main/book/definitions_lorentz.yml"
definitionsFile = wget.download(url)
with open("definitions_lorentz.yml") as file:
# The FullLoader parameter handles the conversion from YAML
# scalar values to Python the dictionary format
definitions = yaml.load(file, Loader=yaml.FullLoader)
features = definitions["features"]
spectators = definitions["spectators"]
labels = definitions["labels"]
nfeatures = definitions["nfeatures"]
nspectators = definitions["nspectators"]
nlabels = definitions["nlabels"]
ntracks = definitions["ntracks"]
Graph datasets#
Here we have to define the graph dataset. We do this in a separate class following this example: https://pytorch-geometric.readthedocs.io/en/latest/notes/create_dataset.html#creating-larger-datasets
Formally, a graph is represented by a triplet \(\mathcal G = (\mathbf{u}, V, E)\), consisting of a graph-level, or global, feature vector \(\mathbf{u}\), a set of \(N^v\) nodes \(V\), and a set of \(N^e\) edges \(E\). The nodes are given by \(V = \{\mathbf{v}_i\}_{i=1:N^v}\), where \(\mathbf{v}_i\) represents the \(i\)th node’s attributes. The edges connect pairs of nodes and are given by \(E = \{\left(\mathbf{e}_k, r_k, s_k\right)\}_{k=1:N^e}\), where \(\mathbf{e}_k\) represents the \(k\)th edge’s attributes, and \(r_k\) and \(s_k\) are the indices of the “receiver” and “sender” nodes, respectively, connected by the \(k\)th edge (from the sender node to the receiver node). The receiver and sender index vectors are an alternative way of encoding the directed adjacency matrix.

# If in colab
if not os.path.exists("LorentzGraphDataset.py"):
urlDSD = "https://raw.githubusercontent.com/jmduarte/iaifi-summer-school/main/book/LorentzGraphDataset.py"
DSD = wget.download(urlDSD)
if not os.path.exists("utils.py"):
urlUtils = "https://raw.githubusercontent.com/jmduarte/iaifi-summer-school/main/book/utils.py"
utils = wget.download(urlUtils)
from LorentzGraphDataset import LorentzGraphDataset
# For Colab
if not os.path.exists("ntuple_merged_90.root"):
urlFILE = "http://opendata.cern.ch/eos/opendata/cms/datascience/HiggsToBBNtupleProducerTool/HiggsToBBNTuple_HiggsToBB_QCD_RunII_13TeV_MC/train/ntuple_merged_90.root"
dataFILE = wget.download(urlFILE)
file_names = ["ntuple_merged_90.root"]
##If you pulled github locally
# if local:
# file_names = [
# "/teams/DSC180A_FA20_A00/b06particlephysics/train/ntuple_merged_10.root"
# ]
# file_names_test = [
# "/teams/DSC180A_FA20_A00/b06particlephysics/test/ntuple_merged_0.root"
# ]
# else:
# file_names = [
# "root://eospublic.cern.ch//eos/opendata/cms/datascience/HiggsToBBNtupleProducerTool/HiggsToBBNTuple_HiggsToBB_QCD_RunII_13TeV_MC/train/ntuple_merged_10.root"
# ]
# file_names_test = [
# "root://eospublic.cern.ch//eos/opendata/cms/datascience/HiggsToBBNtupleProducerTool/HiggsToBBNTuple_HiggsToBB_QCD_RunII_13TeV_MC/test/ntuple_merged_0.root"
# ]
graph_dataset = LorentzGraphDataset(
"ldata_train",
features,
labels,
spectators,
start_event=0,
stop_event=8000,
n_events_merge=1,
file_names=file_names,
)
test_dataset = LorentzGraphDataset(
"ldata_test",
features,
labels,
spectators,
start_event=8001,
stop_event=10001,
n_events_merge=1,
file_names=file_names,
)
print(test_dataset.features)
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
/tmp/ipykernel_5308/1199474778.py in <module>
13 if not os.path.exists("ntuple_merged_90.root"):
14 urlFILE = "http://opendata.cern.ch/eos/opendata/cms/datascience/HiggsToBBNtupleProducerTool/HiggsToBBNTuple_HiggsToBB_QCD_RunII_13TeV_MC/train/ntuple_merged_90.root"
---> 15 dataFILE = wget.download(urlFILE)
16 file_names = ["ntuple_merged_90.root"]
17
/opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages/wget.py in download(url, out, bar)
524 else:
525 binurl = url
--> 526 (tmpfile, headers) = ulib.urlretrieve(binurl, tmpfile, callback)
527 filename = detect_filename(url, out, headers)
528 if outdir:
/opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/urllib/request.py in urlretrieve(url, filename, reporthook, data)
274
275 while True:
--> 276 block = fp.read(bs)
277 if not block:
278 break
/opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/http/client.py in read(self, amt)
463 # Amount is given, implement using readinto
464 b = bytearray(amt)
--> 465 n = self.readinto(b)
466 return memoryview(b)[:n].tobytes()
467 else:
/opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/http/client.py in readinto(self, b)
507 # connection, and the user is reading more bytes than will be provided
508 # (for example, reading in 1k chunks)
--> 509 n = self.fp.readinto(b)
510 if not n and b:
511 # Ideally, we would raise IncompleteRead if the content-length
/opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/socket.py in readinto(self, b)
587 while True:
588 try:
--> 589 return self._sock.recv_into(b)
590 except timeout:
591 self._timeout_occurred = True
KeyboardInterrupt:
Graph neural network#
Here, we recapitulate the “graph network” (GN) formalism [3], which generalizes various GNNs and other similar methods. GNs are graph-to-graph mappings, whose output graphs have the same node and edge structure as the input. Formally, a GN block contains three “update” functions, \(\phi\), and three “aggregation” functions, \(\rho\). The stages of processing in a single GN block are:
\( \begin{align} \mathbf{e}'_k &= \phi^e\left(\mathbf{e}_k, \mathbf{v}_{r_k}, \mathbf{v}_{s_k}, \mathbf{u} \right) & \mathbf{\bar{e}}'_i &= \rho^{e \rightarrow v}\left(E'_i\right) & \text{(Edge block),}\\ \mathbf{v}'_i &= \phi^v\left(\mathbf{\bar{e}}'_i, \mathbf{v}_i, \mathbf{u}\right) & \mathbf{\bar{e}}' &= \rho^{e \rightarrow u}\left(E'\right) & \text{(Node block),}\\ \mathbf{u}' &= \phi^u\left(\mathbf{\bar{e}}', \mathbf{\bar{v}}', \mathbf{u}\right) & \mathbf{\bar{v}}' &= \rho^{v \rightarrow u}\left(V'\right) &\text{(Global block).} \label{eq:gn-functions} \end{align} \)
where \(E'_i = \left\{\left(\mathbf{e}'_k, r_k, s_k \right)\right\}_{r_k=i,\; k=1:N^e}\) contains the updated edge features for edges whose receiver node is the \(i\)th node, \(E' = \bigcup_i E_i' = \left\{\left(\mathbf{e}'_k, r_k, s_k \right)\right\}_{k=1:N^e}\) is the set of updated edges, and \(V'=\left\{\mathbf{v}'_i\right\}_{i=1:N^v}\) is the set of updated nodes.

We will define an interaction network model similar to Ref. [4], but just modeling the particle-particle interactions. It will take as input all of the tracks (with 48 features) without truncating or zero-padding. Another modification is the use of batch normalization [5] layers to improve the stability of the training.
import torch.nn as nn
import torch.nn.functional as F
import torch_geometric.transforms as T
from torch_geometric.nn import EdgeConv, global_mean_pool
from torch.nn import Sequential as Seq, Linear as Lin, ReLU, BatchNorm1d
from torch_scatter import scatter_mean
from torch_geometric.nn import MetaLayer
hidden = 16
outputs = 2
class LorentzEdgeBlock(torch.nn.Module):
def __init__(self):
super(LorentzEdgeBlock, self).__init__()
self.edge_mlp = Seq(Lin(4, hidden), ReLU(), Lin(hidden, hidden))
self.minkowski = torch.from_numpy(
np.array(
[
[-1.0, 0.0, 0.0, 0.0],
[0.0, 1.0, 0.0, 0.0],
[0.0, 0.0, 1.0, 0.0],
[0.0, 0.0, 0.0, 1.0],
],
dtype=np.float32,
)
)
def psi(self, x):
return torch.sign(x) * torch.log(torch.abs(x) + 1)
def innerprod(self, x1, x2):
return torch.sum(
torch.mul(torch.matmul(x1, self.minkowski), x2), 1, keepdim=True
)
def forward(self, src, dest, edge_attr, u, batch):
out = torch.cat(
[
self.innerprod(src, src),
self.innerprod(src, dest),
self.psi(self.innerprod(dest, dest)),
self.psi(self.innerprod(src - dest, src - dest)),
],
dim=1,
)
return self.edge_mlp(out)
class LorentzNodeBlock(torch.nn.Module):
def __init__(self):
super(LorentzNodeBlock, self).__init__()
self.node_mlp_1 = Seq(Lin(1 + hidden, hidden), ReLU(), Lin(hidden, hidden))
self.node_mlp_2 = Seq(Lin(1 + hidden, hidden), ReLU(), Lin(hidden, hidden))
self.minkowski = torch.from_numpy(
np.array(
[
[-1.0, 0.0, 0.0, 0.0],
[0.0, 1.0, 0.0, 0.0],
[0.0, 0.0, 1.0, 0.0],
[0.0, 0.0, 0.0, 1.0],
],
dtype=np.float32,
)
)
def innerprod(self, x1, x2):
return torch.sum(
torch.mul(torch.matmul(x1, self.minkowski), x2), 1, keepdim=True
)
def forward(self, x, edge_index, edge_attr, u, batch):
row, col = edge_index
out = torch.cat([self.innerprod(x[row], x[row]), edge_attr], dim=1)
out = self.node_mlp_1(out)
out = scatter_mean(out, col, dim=0, dim_size=x.size(0))
out = torch.cat([self.innerprod(x, x), out], dim=1)
return self.node_mlp_2(out)
class GlobalBlock(torch.nn.Module):
def __init__(self):
super(GlobalBlock, self).__init__()
self.global_mlp = Seq(Lin(hidden, hidden), ReLU(), Lin(hidden, outputs))
def forward(self, x, edge_index, edge_attr, u, batch):
out = scatter_mean(x, batch, dim=0)
return self.global_mlp(out)
class LorentzInteractionNetwork(torch.nn.Module):
def __init__(self):
super(LorentzInteractionNetwork, self).__init__()
self.lorentzinteractionnetwork = MetaLayer(
LorentzEdgeBlock(), LorentzNodeBlock(), GlobalBlock()
)
def forward(self, x, edge_index, batch):
x, edge_attr, u = self.lorentzinteractionnetwork(
x, edge_index, None, None, batch
)
return u
model = LorentzInteractionNetwork().to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=1e-6)
Define training loop#
@torch.no_grad()
def test(model, loader, total, batch_size, leave=False):
model.eval()
xentropy = nn.CrossEntropyLoss(reduction="mean")
sum_loss = 0.0
t = tqdm(enumerate(loader), total=total / batch_size, leave=leave)
for i, data in t:
data = data.to(device)
y = torch.argmax(data.y, dim=1)
batch_output = model(data.x, data.edge_index, data.batch)
batch_loss_item = xentropy(batch_output, y).item()
sum_loss += batch_loss_item
t.set_description("loss = %.5f" % (batch_loss_item))
t.refresh() # to show immediately the update
return sum_loss / (i + 1)
def train(model, optimizer, loader, total, batch_size, leave=False):
model.train()
xentropy = nn.CrossEntropyLoss(reduction="mean")
sum_loss = 0.0
t = tqdm(enumerate(loader), total=total / batch_size, leave=leave)
for i, data in t:
data = data.to(device)
y = torch.argmax(data.y, dim=1)
optimizer.zero_grad()
batch_output = model(data.x, data.edge_index, data.batch)
batch_loss = xentropy(batch_output, y)
batch_loss.backward()
batch_loss_item = batch_loss.item()
t.set_description("loss = %.5f" % batch_loss_item)
t.refresh() # to show immediately the update
sum_loss += batch_loss_item
optimizer.step()
return sum_loss / (i + 1)
Define training, validation, testing data generators#
from torch_geometric.data import Data, DataListLoader, Batch
from torch.utils.data import random_split
def collate(items):
l = sum(items, [])
return Batch.from_data_list(l)
torch.manual_seed(0)
valid_frac = 0.20
full_length = len(graph_dataset)
valid_num = int(valid_frac * full_length)
batch_size = 32
train_dataset, valid_dataset = random_split(
graph_dataset, [full_length - valid_num, valid_num]
)
train_loader = DataListLoader(
train_dataset, batch_size=batch_size, pin_memory=True, shuffle=True
)
train_loader.collate_fn = collate
valid_loader = DataListLoader(
valid_dataset, batch_size=batch_size, pin_memory=True, shuffle=False
)
valid_loader.collate_fn = collate
test_loader = DataListLoader(
test_dataset, batch_size=batch_size, pin_memory=True, shuffle=False
)
test_loader.collate_fn = collate
train_samples = len(train_dataset)
valid_samples = len(valid_dataset)
test_samples = len(test_dataset)
print(full_length)
print(train_samples)
print(valid_samples)
print(test_samples)
7484
5988
1496
1887
/Users/wmccorma/miniconda3/envs/ml-iaifi/lib/python3.9/site-packages/torch_geometric/deprecation.py:12: UserWarning: 'data.DataListLoader' is deprecated, use 'loader.DataListLoader' instead
warnings.warn(out)
Train#
import os.path as osp
n_epochs = 2
stale_epochs = 0
best_valid_loss = 99999
patience = 5
t = tqdm(range(0, n_epochs))
for epoch in t:
loss = train(
model,
optimizer,
train_loader,
train_samples,
batch_size,
leave=bool(epoch == n_epochs - 1),
)
valid_loss = test(
model,
valid_loader,
valid_samples,
batch_size,
leave=bool(epoch == n_epochs - 1),
)
print("Epoch: {:02d}, Training Loss: {:.4f}".format(epoch, loss))
print(" Validation Loss: {:.4f}".format(valid_loss))
if valid_loss < best_valid_loss:
best_valid_loss = valid_loss
modpath = osp.join("lorentznetwork_best.pth")
print("New best model saved to:", modpath)
torch.save(model.state_dict(), modpath)
stale_epochs = 0
else:
print("Stale epoch")
stale_epochs += 1
if stale_epochs >= patience:
print("Early stopping after %i stale epochs" % patience)
break
Epoch: 00, Training Loss: 0.7810
Validation Loss: 0.7829
New best model saved to: lorentznetwork_best.pth
Epoch: 01, Training Loss: 0.7805
Validation Loss: 0.7821
New best model saved to: lorentznetwork_best.pth
Evaluate on testing data#
# In case you need to load the model from a pth file
# Trained on 4 vectors (as above in notebook)
# urlPTH = "https://raw.githubusercontent.com/jmduarte/iaifi-summer-school/main/book/lorentznetwork_best.pth"
# pthFile = wget.download(urlPTH)
# model.load_state_dict(torch.load("lorentznetwork_best.pth"))
model.eval()
t = tqdm(enumerate(test_loader), total=test_samples / batch_size)
y_test = []
y_predict = []
track_pt = []
for i, data in t:
data = data.to(device)
batchmask = torch.cat([-1.0 * torch.ones(1), data.batch[:-1]], dim=0)
track_pt.append(data.x[batchmask != data.batch, 0])
batch_output = model(data.x, data.edge_index, data.batch)
y_predict.append(batch_output.detach().cpu().numpy())
y_test.append(data.y.cpu().numpy())
track_pt = np.concatenate(track_pt)
y_test = np.concatenate(y_test)
y_predict = np.concatenate(y_predict)
from sklearn.metrics import roc_curve, auc
import matplotlib.pyplot as plt
import mplhep as hep
plt.style.use(hep.style.ROOT)
# create ROC curves
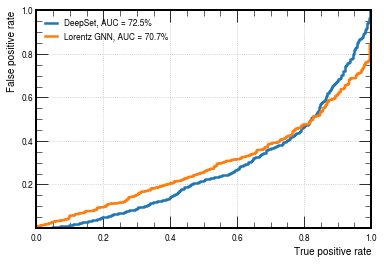
fpr_lorentz, tpr_lorentz, threshold_lorentz = roc_curve(y_test[:, 1], -y_predict[:, 1])
with open("lorentz_roc.npy", "wb") as f:
np.save(f, fpr_lorentz)
np.save(f, tpr_lorentz)
np.save(f, threshold_lorentz)
For colab:
if not os.path.exists("deepset_roc.py"):
urlROC = "https://raw.githubusercontent.com/jmduarte/iaifi-summer-school/main/book/deepset_roc.npy"
rocFile = wget.download(urlROC)
with open("deepset_roc.npy", "rb") as f:
fpr_deepset = np.load(f)
tpr_deepset = np.load(f)
threshold_deepset = np.load(f)
# plot ROC curves
plt.figure()
plt.plot(
tpr_deepset,
fpr_deepset,
lw=2.5,
label="DeepSet, AUC = {:.1f}%".format(auc(fpr_deepset, tpr_deepset) * 100),
)
plt.plot(
tpr_lorentz,
fpr_lorentz,
lw=2.5,
label="Lorentz GNN, AUC = {:.1f}%".format(auc(fpr_lorentz, tpr_lorentz) * 100),
)
plt.xlabel(r"True positive rate")
plt.ylabel(r"False positive rate")
plt.ylim(0.001, 1)
plt.xlim(0, 1)
plt.grid(True)
plt.legend(loc="upper left")
plt.show()
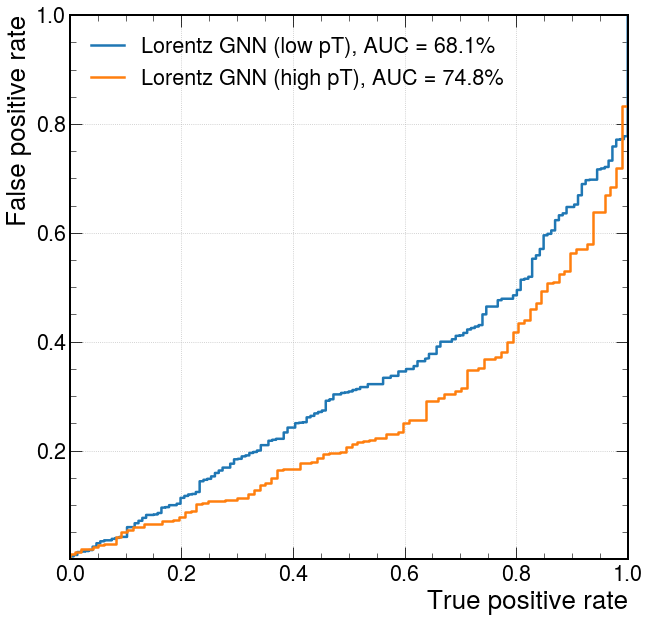
from sklearn.metrics import roc_curve, auc
import matplotlib.pyplot as plt
import mplhep as hep
plt.style.use(hep.style.ROOT)
# create ROC curves
pt_split = 5.0
fpr_lorentz_lowb, tpr_lorentz_lowb, threshold_lorentz_lowb = roc_curve(
y_test[track_pt <= pt_split, 1], -y_predict[track_pt <= pt_split, 1]
)
fpr_lorentz_highb, tpr_lorentz_highb, threshold_lorentz_highb = roc_curve(
y_test[track_pt > pt_split, 1], -y_predict[track_pt > pt_split, 1]
)
# plot ROC curves
plt.figure()
plt.plot(
tpr_lorentz_lowb,
fpr_lorentz_lowb,
lw=2.5,
label="Lorentz GNN (low pT), AUC = {:.1f}%".format(
auc(fpr_lorentz_lowb, tpr_lorentz_lowb) * 100
),
)
plt.plot(
tpr_lorentz_highb,
fpr_lorentz_highb,
lw=2.5,
label="Lorentz GNN (high pT), AUC = {:.1f}%".format(
auc(fpr_lorentz_highb, tpr_lorentz_highb) * 100
),
)
plt.xlabel(r"True positive rate")
plt.ylabel(r"False positive rate")
plt.ylim(0.001, 1)
plt.xlim(0, 1)
plt.grid(True)
plt.legend(loc="upper left")
plt.show()