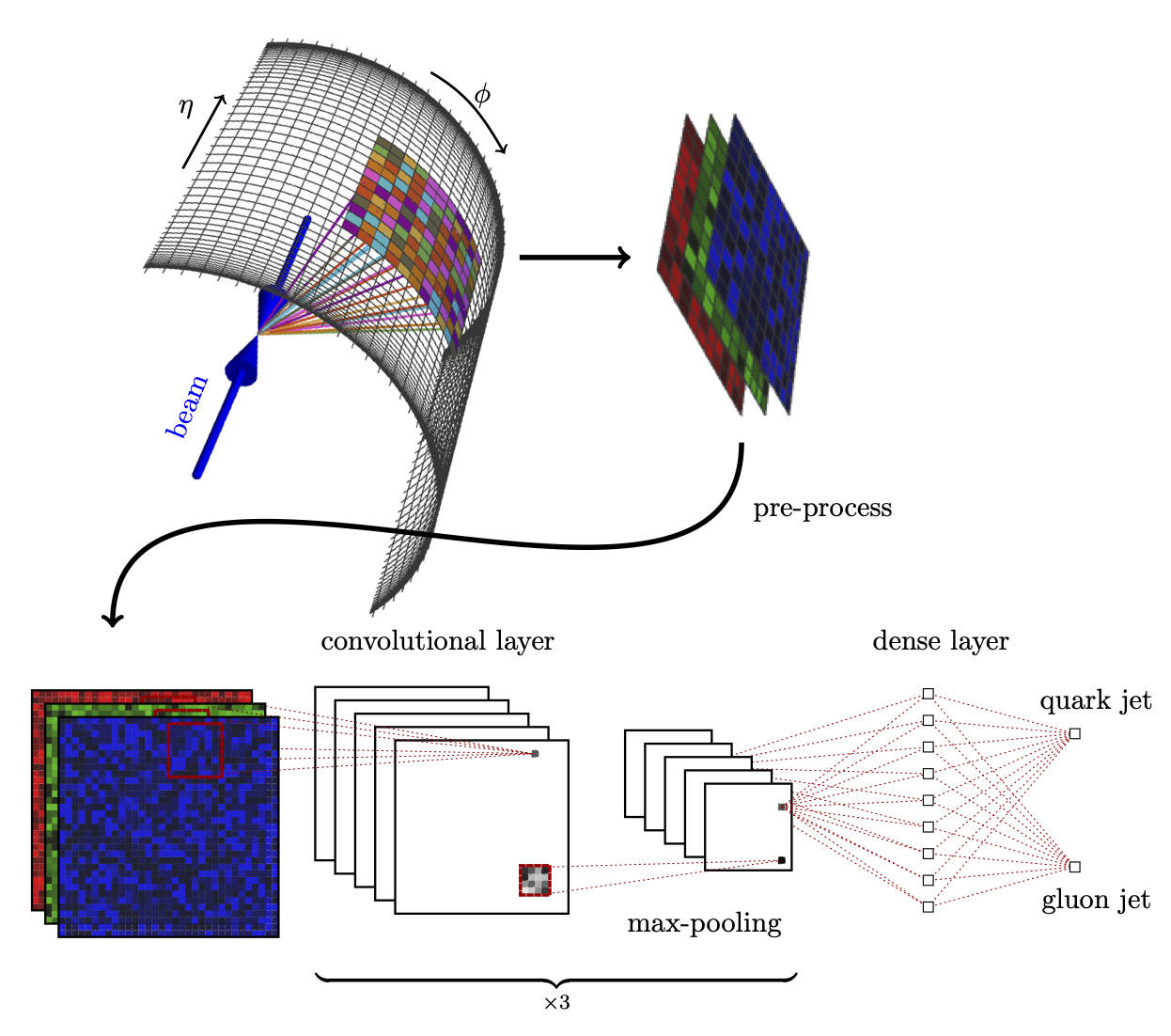
Hands-on 07: Autoencoders for anomaly detection#
This week, we will look at autoencoders for anomaly detection.
The goal is to train an autoencoder to reconstruct QCD (background) jets, which are plentiful at the LHC. Then we will apply it to top quark (signal) jets to see if the reconstruction error is larger. The reconstruction error can then be used as an anomaly score in real data.
This autoencoder architecture used is inspired by this paper: https://arxiv.org/abs/1808.08992.
You may need to install the JetNet library if you don’t already have it.
pip install --user jetnet tables==3.8.0
Download the dataset#
We will use a validation dataset of 400k jets, which is plenty for our purposes. The full dataset is available at https://doi.org/10.5281/zenodo.2603255.
import jetnet
im_size = 16
jet_r = 0.8
max_jets = 50000
# download the validation data (400k jets, which is plenty for our purposes)
# full dataset is available here: https://doi.org/10.5281/zenodo.2603255
data = jetnet.datasets.TopTagging(
jet_type="all",
particle_features=["E", "px", "py", "pz"],
jet_features=["type"],
split="valid",
data_dir="data/",
particle_transform=jetnet.utils.cartesian_to_relEtaPhiPt,
download=True,
)
Downloading val dataset to data/val.h5
Downloading dataset
[..................................................] 0%
[..................................................] 1%
[..................................................] 1%
[..................................................] 1%
[..................................................] 2%
[..................................................] 2%
[█.................................................] 2%
[█.................................................] 2%
[█.................................................] 3%
[█.................................................] 3%
[█.................................................] 3%
[█.................................................] 4%
[█.................................................] 4%
[██................................................] 4%
[██................................................] 5%
[██................................................] 5%
[██................................................] 5%
[██................................................] 5%
[██................................................] 6%
[███...............................................] 6%
[███...............................................] 6%
[███...............................................] 7%
[███...............................................] 7%
[███...............................................] 7%
[███...............................................] 8%
[███...............................................] 8%
[████..............................................] 8%
[████..............................................] 8%
[████..............................................] 9%
[████..............................................] 9%
[████..............................................] 9%
[████..............................................] 10%
[████..............................................] 10%
[█████.............................................] 10%
[█████.............................................] 11%
[█████.............................................] 11%
[█████.............................................] 11%
[█████.............................................] 11%
[█████.............................................] 12%
[██████............................................] 12%
[██████............................................] 12%
[██████............................................] 13%
[██████............................................] 13%
[██████............................................] 13%
[██████............................................] 14%
[██████............................................] 14%
[███████...........................................] 14%
[███████...........................................] 14%
[███████...........................................] 15%
[███████...........................................] 15%
[███████...........................................] 15%
[███████...........................................] 16%
[███████...........................................] 16%
[████████..........................................] 16%
[████████..........................................] 17%
[████████..........................................] 17%
[████████..........................................] 17%
[████████..........................................] 18%
[████████..........................................] 18%
[█████████.........................................] 18%
[█████████.........................................] 18%
[█████████.........................................] 19%
[█████████.........................................] 19%
[█████████.........................................] 19%
[█████████.........................................] 20%
[█████████.........................................] 20%
[██████████........................................] 20%
[██████████........................................] 21%
[██████████........................................] 21%
[██████████........................................] 21%
[██████████........................................] 21%
[██████████........................................] 22%
[███████████.......................................] 22%
[███████████.......................................] 22%
[███████████.......................................] 23%
[███████████.......................................] 23%
[███████████.......................................] 23%
[███████████.......................................] 24%
[███████████.......................................] 24%
[████████████......................................] 24%
[████████████......................................] 24%
[████████████......................................] 25%
[████████████......................................] 25%
[████████████......................................] 25%
[████████████......................................] 26%
[████████████......................................] 26%
[█████████████.....................................] 26%
[█████████████.....................................] 27%
[█████████████.....................................] 27%
[█████████████.....................................] 27%
[█████████████.....................................] 27%
[█████████████.....................................] 28%
[██████████████....................................] 28%
[██████████████....................................] 28%
[██████████████....................................] 29%
[██████████████....................................] 29%
[██████████████....................................] 29%
[██████████████....................................] 30%
[██████████████....................................] 30%
[███████████████...................................] 30%
[███████████████...................................] 30%
[███████████████...................................] 31%
[███████████████...................................] 31%
[███████████████...................................] 31%
[███████████████...................................] 32%
[███████████████...................................] 32%
[████████████████..................................] 32%
[████████████████..................................] 33%
[████████████████..................................] 33%
[████████████████..................................] 33%
[████████████████..................................] 34%
[████████████████..................................] 34%
[█████████████████.................................] 34%
[█████████████████.................................] 34%
[█████████████████.................................] 35%
[█████████████████.................................] 35%
[█████████████████.................................] 35%
[█████████████████.................................] 36%
[█████████████████.................................] 36%
[██████████████████................................] 36%
[██████████████████................................] 37%
[██████████████████................................] 37%
[██████████████████................................] 37%
[██████████████████................................] 37%
[██████████████████................................] 38%
[███████████████████...............................] 38%
[███████████████████...............................] 38%
[███████████████████...............................] 39%
[███████████████████...............................] 39%
[███████████████████...............................] 39%
[███████████████████...............................] 40%
[███████████████████...............................] 40%
[████████████████████..............................] 40%
[████████████████████..............................] 40%
[████████████████████..............................] 41%
[████████████████████..............................] 41%
[████████████████████..............................] 41%
[████████████████████..............................] 42%
[████████████████████..............................] 42%
[█████████████████████.............................] 42%
[█████████████████████.............................] 43%
[█████████████████████.............................] 43%
[█████████████████████.............................] 43%
[█████████████████████.............................] 43%
[█████████████████████.............................] 44%
[██████████████████████............................] 44%
[██████████████████████............................] 44%
[██████████████████████............................] 45%
[██████████████████████............................] 45%
[██████████████████████............................] 45%
[██████████████████████............................] 46%
[██████████████████████............................] 46%
[███████████████████████...........................] 46%
[███████████████████████...........................] 46%
[███████████████████████...........................] 47%
[███████████████████████...........................] 47%
[███████████████████████...........................] 47%
[███████████████████████...........................] 48%
[███████████████████████...........................] 48%
[████████████████████████..........................] 48%
[████████████████████████..........................] 49%
[████████████████████████..........................] 49%
[████████████████████████..........................] 49%
[████████████████████████..........................] 50%
[████████████████████████..........................] 50%
[█████████████████████████.........................] 50%
[█████████████████████████.........................] 50%
[█████████████████████████.........................] 51%
[█████████████████████████.........................] 51%
[█████████████████████████.........................] 51%
[█████████████████████████.........................] 52%
[█████████████████████████.........................] 52%
[██████████████████████████........................] 52%
[██████████████████████████........................] 53%
[██████████████████████████........................] 53%
[██████████████████████████........................] 53%
[██████████████████████████........................] 53%
[██████████████████████████........................] 54%
[███████████████████████████.......................] 54%
[███████████████████████████.......................] 54%
[███████████████████████████.......................] 55%
[███████████████████████████.......................] 55%
[███████████████████████████.......................] 55%
[███████████████████████████.......................] 56%
[███████████████████████████.......................] 56%
[████████████████████████████......................] 56%
[████████████████████████████......................] 56%
[████████████████████████████......................] 57%
[████████████████████████████......................] 57%
[████████████████████████████......................] 57%
[████████████████████████████......................] 58%
[████████████████████████████......................] 58%
[█████████████████████████████.....................] 58%
[█████████████████████████████.....................] 59%
[█████████████████████████████.....................] 59%
[█████████████████████████████.....................] 59%
[█████████████████████████████.....................] 59%
[█████████████████████████████.....................] 60%
[██████████████████████████████....................] 60%
[██████████████████████████████....................] 60%
[██████████████████████████████....................] 61%
[██████████████████████████████....................] 61%
[██████████████████████████████....................] 61%
[██████████████████████████████....................] 62%
[██████████████████████████████....................] 62%
[███████████████████████████████...................] 62%
[███████████████████████████████...................] 62%
[███████████████████████████████...................] 63%
[███████████████████████████████...................] 63%
[███████████████████████████████...................] 63%
[███████████████████████████████...................] 64%
[███████████████████████████████...................] 64%
[████████████████████████████████..................] 64%
[████████████████████████████████..................] 65%
[████████████████████████████████..................] 65%
[████████████████████████████████..................] 65%
[████████████████████████████████..................] 66%
[████████████████████████████████..................] 66%
[█████████████████████████████████.................] 66%
[█████████████████████████████████.................] 66%
[█████████████████████████████████.................] 67%
[█████████████████████████████████.................] 67%
[█████████████████████████████████.................] 67%
[█████████████████████████████████.................] 68%
[█████████████████████████████████.................] 68%
[██████████████████████████████████................] 68%
[██████████████████████████████████................] 69%
[██████████████████████████████████................] 69%
[██████████████████████████████████................] 69%
[██████████████████████████████████................] 69%
[██████████████████████████████████................] 70%
[███████████████████████████████████...............] 70%
[███████████████████████████████████...............] 70%
[███████████████████████████████████...............] 71%
[███████████████████████████████████...............] 71%
[███████████████████████████████████...............] 71%
[███████████████████████████████████...............] 72%
[███████████████████████████████████...............] 72%
[████████████████████████████████████..............] 72%
[████████████████████████████████████..............] 72%
[████████████████████████████████████..............] 73%
[████████████████████████████████████..............] 73%
[████████████████████████████████████..............] 73%
[████████████████████████████████████..............] 74%
[████████████████████████████████████..............] 74%
[█████████████████████████████████████.............] 74%
[█████████████████████████████████████.............] 75%
[█████████████████████████████████████.............] 75%
[█████████████████████████████████████.............] 75%
[█████████████████████████████████████.............] 75%
[█████████████████████████████████████.............] 76%
[██████████████████████████████████████............] 76%
[██████████████████████████████████████............] 76%
[██████████████████████████████████████............] 77%
[██████████████████████████████████████............] 77%
[██████████████████████████████████████............] 77%
[██████████████████████████████████████............] 78%
[██████████████████████████████████████............] 78%
[███████████████████████████████████████...........] 78%
[███████████████████████████████████████...........] 78%
[███████████████████████████████████████...........] 79%
[███████████████████████████████████████...........] 79%
[███████████████████████████████████████...........] 79%
[███████████████████████████████████████...........] 80%
[███████████████████████████████████████...........] 80%
[████████████████████████████████████████..........] 80%
[████████████████████████████████████████..........] 81%
[████████████████████████████████████████..........] 81%
[████████████████████████████████████████..........] 81%
[████████████████████████████████████████..........] 82%
[████████████████████████████████████████..........] 82%
[█████████████████████████████████████████.........] 82%
[█████████████████████████████████████████.........] 82%
[█████████████████████████████████████████.........] 83%
[█████████████████████████████████████████.........] 83%
[█████████████████████████████████████████.........] 83%
[█████████████████████████████████████████.........] 84%
[█████████████████████████████████████████.........] 84%
[██████████████████████████████████████████........] 84%
[██████████████████████████████████████████........] 85%
[██████████████████████████████████████████........] 85%
[██████████████████████████████████████████........] 85%
[██████████████████████████████████████████........] 85%
[██████████████████████████████████████████........] 86%
[███████████████████████████████████████████.......] 86%
[███████████████████████████████████████████.......] 86%
[███████████████████████████████████████████.......] 87%
[███████████████████████████████████████████.......] 87%
[███████████████████████████████████████████.......] 87%
[███████████████████████████████████████████.......] 88%
[███████████████████████████████████████████.......] 88%
[████████████████████████████████████████████......] 88%
[████████████████████████████████████████████......] 88%
[████████████████████████████████████████████......] 89%
[████████████████████████████████████████████......] 89%
[████████████████████████████████████████████......] 89%
[████████████████████████████████████████████......] 90%
[████████████████████████████████████████████......] 90%
[█████████████████████████████████████████████.....] 90%
[█████████████████████████████████████████████.....] 91%
[█████████████████████████████████████████████.....] 91%
[█████████████████████████████████████████████.....] 91%
[█████████████████████████████████████████████.....] 91%
[█████████████████████████████████████████████.....] 92%
[██████████████████████████████████████████████....] 92%
[██████████████████████████████████████████████....] 92%
[██████████████████████████████████████████████....] 93%
[██████████████████████████████████████████████....] 93%
[██████████████████████████████████████████████....] 93%
[██████████████████████████████████████████████....] 94%
[██████████████████████████████████████████████....] 94%
[███████████████████████████████████████████████...] 94%
[███████████████████████████████████████████████...] 94%
[███████████████████████████████████████████████...] 95%
[███████████████████████████████████████████████...] 95%
[███████████████████████████████████████████████...] 95%
[███████████████████████████████████████████████...] 96%
[███████████████████████████████████████████████...] 96%
[████████████████████████████████████████████████..] 96%
[████████████████████████████████████████████████..] 97%
[████████████████████████████████████████████████..] 97%
[████████████████████████████████████████████████..] 97%
[████████████████████████████████████████████████..] 97%
[████████████████████████████████████████████████..] 98%
[█████████████████████████████████████████████████.] 98%
[█████████████████████████████████████████████████.] 98%
[█████████████████████████████████████████████████.] 99%
[█████████████████████████████████████████████████.] 99%
[█████████████████████████████████████████████████.] 99%
[█████████████████████████████████████████████████.] 100%
[█████████████████████████████████████████████████.] 100%
[██████████████████████████████████████████████████] 100%
Transform and split the data#
The data is originally up to 200 particles per jet (zero-padded), and the features are the standard 4-vectors \((E, p_x, p_y, p_z)\). We can assume the particles are massless so \(E=\sqrt{p_x^2+p_y^2+p_z^2}\) and there are only 3 degrees of freedom.
We will transform to relative coordinates centered on the jet using the function jetnet.utils.cartesian_to_relEtaPhiPt:
\begin{align} \eta^\mathrm{rel} &=\eta^\mathrm{particle} - \eta^\mathrm{jet}\ \phi^\mathrm{rel} &=\phi^\mathrm{particle} - \phi^\mathrm{jet} \pmod{2\pi}\ p_\mathrm{T}^\mathrm{rel} &= p_\mathrm{T}^\mathrm{particle}/p_\mathrm{T}^\mathrm{jet} \end{align}
import numpy as np
indices = np.random.permutation(np.arange(len(data)))[:max_jets]
# transform the data
transformed_particle_data = data.particle_transform(data.particle_data[indices])
# split qcd background and top quark signal
qcd_data = transformed_particle_data[data.jet_data[indices][:, 0] == 0]
top_data = transformed_particle_data[data.jet_data[indices][:, 0] == 1]
from sklearn.model_selection import train_test_split
qcd_train, qcd_test = train_test_split(qcd_data, test_size=0.2, random_state=42)
top_train, top_test = train_test_split(top_data, test_size=0.2, random_state=42)
Convert data to jet images#
import numpy as np
# convert full dataset
qcd_train_images = np.expand_dims(jetnet.utils.to_image(qcd_train, im_size=im_size, maxR=jet_r), axis=-1)
qcd_test_images = np.expand_dims(jetnet.utils.to_image(qcd_test, im_size=im_size, maxR=jet_r), axis=-1)
top_test_images = np.expand_dims(jetnet.utils.to_image(top_test, im_size=im_size, maxR=jet_r), axis=-1)
# rescale so sum is 1 (it should be close already)
qcd_train_images = qcd_train_images / np.sum(qcd_train_images.reshape(-1, 1, 1, 1, im_size * im_size), axis=-1)
qcd_test_images = qcd_test_images / np.sum(qcd_test_images.reshape(-1, 1, 1, 1, im_size * im_size), axis=-1)
top_test_images = top_test_images / np.sum(top_test_images.reshape(-1, 1, 1, 1, im_size * im_size), axis=-1)
Visualize the jet images#
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
def plot_jet_images(images, titles, filename="jet_image.pdf"):
n_images = len(images)
plt.figure(figsize=(5 * n_images, 5))
for i, (image, title) in enumerate(zip(images, titles)):
plt.subplot(1, n_images, i + 1)
plt.title(title)
plt.imshow(image, origin="lower", norm=LogNorm(vmin=1e-3, vmax=1))
cbar = plt.colorbar()
plt.xlabel(r"$\Delta\eta$ cell", fontsize=15)
plt.ylabel(r"$\Delta\phi$ cell", fontsize=15)
cbar.set_label(r"$p_T/p_T^{jet}$", fontsize=15)
plt.tight_layout()
plt.savefig(filename)
plot_jet_images([qcd_test_images[0], top_test_images[0]], ["QCD jet image", "Top quark jet image"])

Define the autoencoder model#
from tensorflow.keras.models import Model
from tensorflow.keras.layers import (
Dense,
Input,
Conv2D,
Conv2DTranspose,
Reshape,
Flatten,
Softmax,
)
x_in = Input(shape=(im_size, im_size, 1))
x = Conv2D(128, kernel_size=(3, 3), strides=(2, 2), activation="relu", padding="same")(x_in)
x = Conv2D(128, kernel_size=(3, 3), strides=(2, 2), activation="relu", padding="same")(x)
x = Flatten()(x)
x_enc = Dense(2, name="bottleneck")(x)
x = Dense(int(im_size * im_size / 16) * 128, activation="relu")(x_enc)
x = Reshape((int(im_size / 4), int(im_size / 4), 128))(x)
x = Conv2DTranspose(128, kernel_size=(3, 3), strides=(2, 2), activation="relu", padding="same")(x)
x = Conv2DTranspose(1, kernel_size=(3, 3), strides=(2, 2), activation="linear", padding="same")(x)
x_out = Softmax(name="softmax", axis=[-2, -3])(x)
model = Model(inputs=x_in, outputs=x_out, name="autoencoder")
model.compile(loss="mse", optimizer="adam")
model.summary()
# save the encoder-only model for easy access to latent space
encoder = Model(inputs=x_in, outputs=x_enc, name="encoder")
Model: "autoencoder"
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┓ ┃ Layer (type) ┃ Output Shape ┃ Param # ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━┩ │ input_layer (InputLayer) │ (None, 16, 16, 1) │ 0 │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ conv2d (Conv2D) │ (None, 8, 8, 128) │ 1,280 │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ conv2d_1 (Conv2D) │ (None, 4, 4, 128) │ 147,584 │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ flatten (Flatten) │ (None, 2048) │ 0 │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ bottleneck (Dense) │ (None, 2) │ 4,098 │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ dense (Dense) │ (None, 2048) │ 6,144 │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ reshape (Reshape) │ (None, 4, 4, 128) │ 0 │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ conv2d_transpose │ (None, 8, 8, 128) │ 147,584 │ │ (Conv2DTranspose) │ │ │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ conv2d_transpose_1 │ (None, 16, 16, 1) │ 1,153 │ │ (Conv2DTranspose) │ │ │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ softmax (Softmax) │ (None, 16, 16, 1) │ 0 │ └─────────────────────────────────┴────────────────────────┴───────────────┘
Total params: 307,843 (1.17 MB)
Trainable params: 307,843 (1.17 MB)
Non-trainable params: 0 (0.00 B)
Train the autoencoder model#
history = model.fit(
qcd_train_images,
qcd_train_images,
batch_size=32,
epochs=10,
verbose=0,
validation_data=(qcd_test_images, qcd_test_images),
)
Reconstruction performance#
qcd_reco_image = model.predict(qcd_test_images[0:1], verbose=0).reshape(im_size, im_size)
plot_jet_images([qcd_test_images[0], qcd_reco_image], ["Input QCD jet image", "Reconstructed QCD jet image"])

top_reco_image = model.predict(top_test_images[0:1], verbose=0).reshape(im_size, im_size)
plot_jet_images([top_test_images[0], top_reco_image], ["Input top quark jet image", "Reconstructed top quark jet image"])

Anomaly detection performance#
qcd_reco_images = model.predict(qcd_test_images, verbose=0)
top_reco_images = model.predict(top_test_images, verbose=0)
diff_qcd = np.power((qcd_reco_images - qcd_test_images), 2)
loss_qcd = np.mean(diff_qcd.reshape(-1, im_size * im_size), axis=-1)
diff_top = np.power((top_reco_images - top_test_images), 2)
loss_top = np.mean(diff_top.reshape(-1, im_size * im_size), axis=-1)
loss_all = np.concatenate([loss_qcd, loss_top])
plt.figure()
bins = np.arange(0, np.max(loss_all), np.max(loss_all) / 100)
plt.hist(loss_qcd, label="QCD jets", bins=bins, alpha=0.8)
plt.hist(loss_top, label="Top quark jets", bins=bins, alpha=0.8)
plt.legend(title="Autoencoder")
plt.xlabel("MSE loss")
plt.ylabel("Jets")
plt.xlim(0, np.max(loss_all))
plt.show()

Exercises#
Plot the ROC curve for the MSE loss of the autoencoder on the merged testing sample of QCD and top quark jets, assuming a label of 1 for top quark jets and a label of 0 for QCD jets. Report the AUC.
Perform a PCA on only the QCD training images using
sklearn.decomposition.PCAwith 2 components. Note you will have to reshape the image tensors so that they are 2D instead of 4D (as required by the autoencoder), e.g.qcd_test_images.reshape(-1, im_size * im_size)). Plot the distribution of the reconstruction losses for top quark jets and QCD jets separately. Hint: review https://rittikghosh.com/autoencoder.html.Plot the PCA ROC curve similar to part 1. Report the AUC.
Plot the 2D latent space for the QCD and top quark test images for both the autoencoder and the PCA.